Two way repeated measures analysis in R
In this tutorial you will learn how to carry out analysis for two way repeated measures anova using R studio. A two way repeated measures anova compares the mean differences between groups that have been split on two within subject factors. These factors are also known as independent variables.
Two way repeated measures anova is also known as:
- Two factor repeated measures anova
- Two factor or two way anova with repeated measures
- Within within subjects anova
A two way repeated measures anova is often used in studies where you have measured:
- A dependent variable over two or more time points
- When subjects have undergone two or more conditions
You should also know how to identify a two way repeated measures anova from mixed anova. Mixed anova is very similar to two way repeated measures anova because both tests involve two factors. In a mixed anova, subjects that undergo each condition are different. Whereas in two way repeated measures anova same subjects undergo both conditions.
Description of data
The data file which is used for analysis contains subject ID, condition, time interval and response variable. Here subject ID represents the subjects or individuals used to record the response variable measurements. These subjects have a condition where male and female individuals were used. The response variable measurements were recorded at a four different time intervals. In this data set you can see the same subjects were repeated to record the response variable at different time intervals rather than using different individuals. As same subjects were repeated, so we shall proceed with two way repeated measures anova in R studio.
Clear R environment
As a first step, I always recommend to clear data objects and values in global environment with rm() function. Set TRUE value for the argument all to remove objects and values if you have created earlier. Shut down all the graphic windows by using graphics.off() function. Putting the value “cls” in shell() function will clear the console environment.
rm(list = ls(all = TRUE))
graphics.off()
shell("cls")Importing data
To import the data set I have placed the CSV data file in the project working directory. Create an object data and assign to it a function which I am calling as read.csv(). In argument file after equal sign within quotations just press tab button to access the files present in the working directory. Now we need to choose the respective CSV data file. In the next argument header type TRUE to indicate that the first row of the data file contains variable names or headings.
In the next line we can use the head() function to print the first six rows of the data frame. Here I am converting the first 2 variables as factor variables by using as.factor() function. Use attach() function for data object to mask the components of the variables in the data frame.
data <- read.csv(file = "data.csv",
header = TRUE)
head(data)
data$group = as.factor(data$group)
data$time = as.factor(data$time)
attach(data)# subID group time resp.var
# 1 1 male day1 26
# 2 2 male day1 27
# 3 3 male day1 24
# 4 4 female day1 18
# 5 5 female day1 17
# 6 6 female day1 16Fitting ANOVA model
Let’s proceed with analysis of variance to see how the response variable changes as the function of condition and time applied to each participant. The aov() function is used to fit analysis of variance model. The function is same as used in two way anova with only difference in error argument. Here you need to define error as ratio between subject ID and interaction of group and time factor. For denominator you can also use group, time and interaction term combined with plus sign. Summary() function for model object will print the results.
model.aov <- aov(resp.var ~
group * time +
Error(subID/(group*time)))
#OR
model.aov <- aov(resp.var ~
group * time +
Error(subID/(group + time + group:time)))
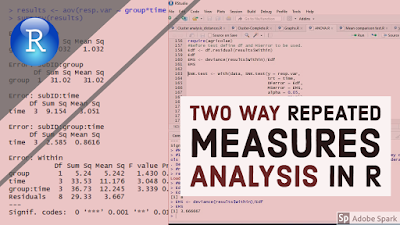
summary(model.aov)#
# Error: subID
# Df Sum Sq Mean Sq
# group 1 408 408
#
# Error: subID:group
# Df Sum Sq Mean Sq
# group 1 67.25 67.25
#
# Error: subID:time
# Df Sum Sq Mean Sq
# time 3 356.7 118.9
#
# Error: subID:group:time
# Df Sum Sq Mean Sq
# time 3 49.94 16.65
#
# Error: Within
# Df Sum Sq Mean Sq F value Pr(>F)
# group 1 13.86 13.863 6.105 0.03866 *
# time 3 52.79 17.596 7.749 0.00943 **
# group:time 3 2.60 0.868 0.382 0.76887
# Residuals 8 18.17 2.271
# ---
# Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The results showed that the error function has decomposed the residuals into:
- Subject ID
- Subject ID and group interaction
- Subject ID and time interaction
- Three way interaction of subject ID, group and time
This decomposition of error term considerably reduce the residual source of variance for the calculation of F value. Both the time and group variables significantly influence the response variable. However, their interaction was non-significant.
Mean comparison test
Now let’s compute mean comparisons to observe the differences between main effects.
SNK test
SNK test is derived from Tukey, but it is less conservative. Tukey test controls the error for all comparisons, where SNK only controls for comparisons under consideration. Before using SNK.test() function, first define error degree of freedom and error mean square value that will be used in this function.
Edf <- df.residual(model.aov$Within)
Edf# [1] 8EMS <- deviance(model.aov$Within)/Edf
EMS# [1] 2.270833To apply SNK test use SNK.test() function. This function requires:
- Y argument that specify the response variable
- TRT argument that represents the treatment factor variable applied to each experimental unit as group and time in this example.
- DF error and MS error represents degree of freedom and mean square for error term as determined earlier
- Alpha shows the significance level. Default value is 5 percent.
- Group argument specify the formation of treatment groups. True value for group will result in treatment groups with lettering. False value for group will provide information regarding comparisons between treatments.
library(agricolae)
SNK.test1 <- SNK.test(y = resp.var,
trt = group,
DFerror = Edf,
MSerror = EMS,
alpha = 0.05,
group = TRUE)
print(SNK.test1)# $statistics
# MSerror Df Mean CV
# 2.270833 8 14.33333 10.51345
#
# $parameters
# test name.t ntr alpha
# SNK group 2 0.05
#
# $snk
# Table CriticalRange
# 2 3.261182 1.418656
#
# $means
# resp.var std r Min Max Q25 Q50 Q75
# female 9.833333 4.687184 12 4 18 6.75 9.0 11.50
# male 18.833333 4.687184 12 13 27 15.75 17.5 21.75
#
# $comparison
# NULL
#
# $groups
# resp.var groups
# male 18.833333 a
# female 9.833333 b
#
# attr(,"class")
# [1] "group"Similarly, for mean comparisons in time variable just replace group with time in trt argument.
SNK.test2 <- SNK.test(y = resp.var,
trt = time,
DFerror = Edf,
MSerror = EMS,
alpha = 0.05,
group = TRUE)
print(SNK.test2)# $statistics
# MSerror Df Mean CV
# 2.270833 8 14.33333 10.51345
#
# $parameters
# test name.t ntr alpha
# SNK time 4 0.05
#
# $snk
# Table CriticalRange
# 2 3.261182 2.006283
# 3 4.041036 2.486050
# 4 4.528810 2.786128
#
# $means
# resp.var std r Min Max Q25 Q50 Q75
# day1 21.333333 4.885352 6 16 27 17.25 21.0 25.50
# day2 13.833333 6.145459 6 7 21 9.00 13.5 18.75
# day3 12.500000 3.937004 6 8 17 9.25 12.5 15.75
# day4 9.666667 5.240865 6 4 16 5.25 9.5 13.75
#
# $comparison
# NULL
#
# $groups
# resp.var groups
# day1 21.333333 a
# day2 13.833333 b
# day3 12.500000 b
# day4 9.666667 c
#
# attr(,"class")
# [1] "group"Visualizing results
Barplot for time variable
You can plot bar graph with standard error at the top of each bar from the output of SNK test applied earlier. The below code shows how the plot will be drawn using ggplot() function.
library(ggplot2)
SE1 = SNK.test1$means$std/sqrt(SNK.test1$means$r)
ggplot(data=SNK.test1$means,
aes(x= rownames(SNK.test1$means),
y= resp.var,
fill=rownames(SNK.test1$means))) +
geom_bar(stat="identity") +
geom_errorbar(aes(ymax = resp.var + std/sqrt(r),
ymin = resp.var - std/sqrt(r)),
position = position_dodge(width=0.9),
width = 0.25) +
labs(title = "",
x = "group",
y = "Response variable",
fill = "group") +
geom_text(data = SNK.test1$groups,
aes(x = rownames(SNK.test1$groups),
y = resp.var + SE1,
label = as.matrix(groups)),
position = position_dodge(width = 0.9),
vjust = -(0.5))Barplot for group variable
SE2 = SNK.test2$means$std/sqrt(SNK.test2$means$r)
ggplot(data=SNK.test2$means,
aes(x= rownames(SNK.test2$means),
y= resp.var,
fill=rownames(SNK.test2$means))) +
geom_bar(stat="identity") +
geom_errorbar(aes(ymax = resp.var + std/sqrt(r),
ymin = resp.var - std/sqrt(r)),
position = position_dodge(width=0.9),
width = 0.25) +
labs(title = "",
x = "time",
y = "Response variable",
fill = "time") +
geom_text(data = SNK.test2$groups,
aes(x = rownames(SNK.test2$groups),
y = resp.var + SE2,
label = as.matrix(groups)),
position = position_dodge(width = 0.9),
vjust = -(0.5))Please comment below if you have any questions.
Download data file — Click_here
Download Rscript — Click_here
Download R program —
Click_hereDownload R studio —
Click_here



For some reason, the "error within" section of the summary(model.aov) is not showing up in my R console. Is there a common reason why?
ReplyDelete